DNA damage & repair
- raghukulchaudhary

- Jul 26, 2018
- 6 min read
Updated: Jul 29, 2018
DNA DAMAGE

DNA damage is an alteration in the chemical structure of DNA, such as a break in a strand of DNA, a base missing from the backbone of DNA, or a chemically changed base.DNA could be damaged from various factors:
Due to environmental factors and normal metabolic processes inside the cell.
Linked with variety of genetically inherited disorders, in aging and in carcinogenesis.
Failure to repair DNA damage may result in blockages of transcription and replication.
Sources of DNA Damage
Endogenous damage: These are generally caused,such as attack by reactive oxygen species produced from normal metabolic byproducts.
Exogenous damage: Usually caused by external agents such as
UV [UV 200-300 nm] radiations, X- rays and gamma rays
human-made mutagenic chemicals (DNA intercalating agents )
cancer chemotherapy and radiotherapy
Types of Damage
The three main types of damage to DNA due to endogenous cellular processes are listed below:
1. Oxidation (loss of electrons) of bases [e.g. 8-oxo-7,8-dihydroguanine (8-oxoG)] and generation of DNA strand interruptions from reactive oxygen species.Oxidative damage results from aerobic metabolism and environmental toxins.Oxidation of Guanine Forms 8-Oxoguanine.Replication of the 8-oxoG strand preferentially mispairs with A.
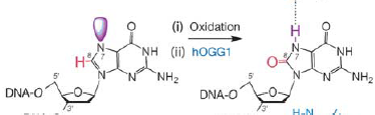
2. Alkylation (transfer of alkyl group) of bases (usually methylation), such as formation of 7-methylguanine, 1-methyladenine, 6 -methylguanine.Many environmental chemicals, including "natural" ones can also modify DNA bases, frequently by addition of a methyl or other alkyl group (alkylation).
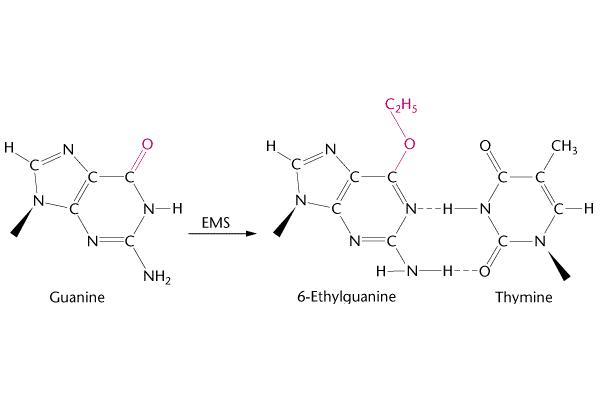
Above we can see how Guanine can change into 6-Ethylquanine under Ethylmethane Sulfonate (EMS). One of the best examples can be taken from Mustard gas which was used in the world war I. Some more examples of Alkylation could be seen in the picture below.
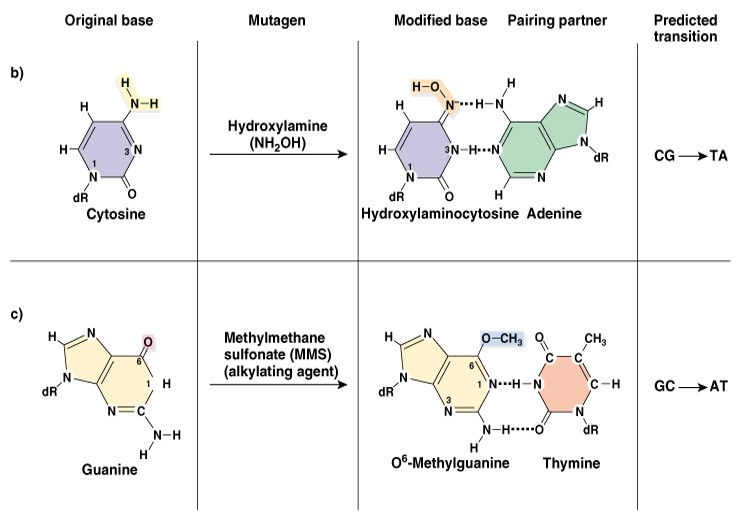
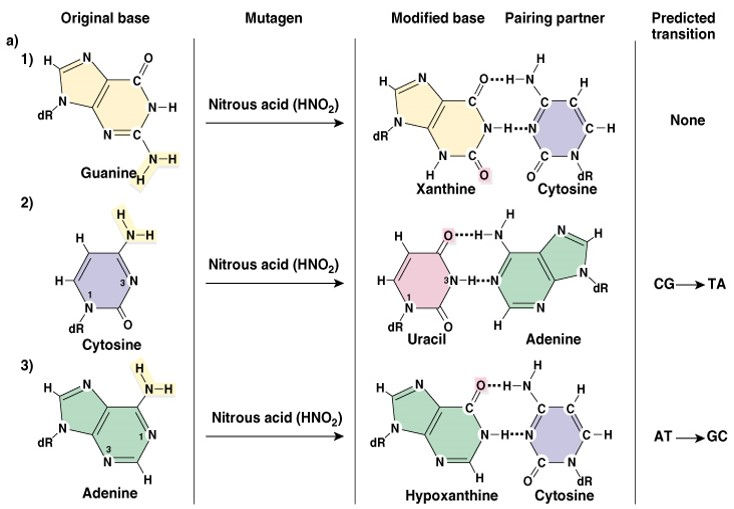
3. Hydrolysis (cleavage of chemical bonds): Hydrolysis of bases usually occurs by deamination, depurination or depyrimidination.
Deamination: Deamination is the removal of an amine group from a molecule. Enzymes that catalyse this reaction are called deaminases.
Oxidative deamination of A,G,C (amino group to keto- group)
Adenine – deamination- hypoxanthine =C
Cytosine – deamination- uracil =A
Guanine – deamination- Xanthine =C
Depurination/Depyrimidination: This can be defined as the removal of purine or pyrimidine bases from DNA.
In Depurination/Depyrimidination the glycosyl bond linking DNA bases with deoxyribose is liable under physiological changes.Loss of a purine or pyrimidine base creates apurinic/apyrimidinic (AP) site (abasic site). Figure of an AP site is shown below
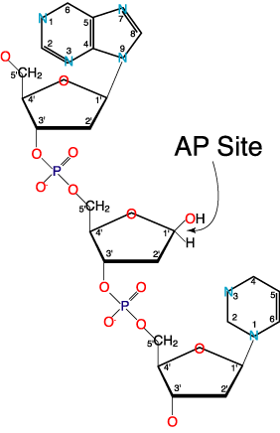
Some more examples are shown in the picture below
DNA REPAIR
DNA Repair - (Biological repair mechanisms of mutations)
Since most mutations are deleterious, DNA repair systems are vital to the survival of all organisms. Living cells contain several DNA repair systems that can fix different type of DNA alterations.
In most cases, DNA repair is a multi-step process:
– An irregularity in DNA structure is detected
– The abnormal DNA is removed
– Normal DNA is synthesized
Common Types of DNA repair systems
1) Direct Repair: An enzymes recognizes an incorrect alternation in DNA structure and directly converts it back to a correct structure.
2) Base excision repair and nucleotide excision repair: An abnormal base or nucleotide is first recognized and removed from DNA. A segment of DNA in this region is excised, and then the complementary DNA strand is used as a template to synthesize a normal DNA strand.
3) Mismatch Repair: Similar to excision repair except that the DNA defect is a base pair mismatch in the DNA,not an abnormal nucleotide. This mismatch is recognised, and a segment of DNA in this region is removed. The parental strand is used to synthesize a normal daughter strand of DNA.
4) Homologous recombination repair: Occurs at double-strand breaks or when DNA causes a gap in synthesis during DNA replication. The strands of a normal sister chromatid are used to repair a damaged sister chromatid.
5) Non-homologous end joining: Occurs at double stranded breaks. The broken ends are recognised by proteins that keep the ends together; the broken ends are eventually rejoined.
Damaged Bases Can Be Directly Repaired
In a few cases, the covalent modifications of nucleotides can be reversed by specific enzymes
1) Photolyase can repair thymine dimers
It splits the dimers restoring the DNA to its original condition
2) O6-alkylguanine alkyltransferase repairs alkylated bases
It transfers the methyl or ethyl group from the base to a cysteine side chain within the alkyltransferase protein
permanently inactivates alkyltransferase

Base Excision Repair:
Base Excision Repair (BER) involves enzymes known as DNA N-glycosylases:
can recognize an abnormal base
cleave the bond between abnormal base and the sugar in the DNA
This repair system can eliminate abnormal bases such as:
Uracil; Thymine dimers
3-methyladenine; 7-methylguanine
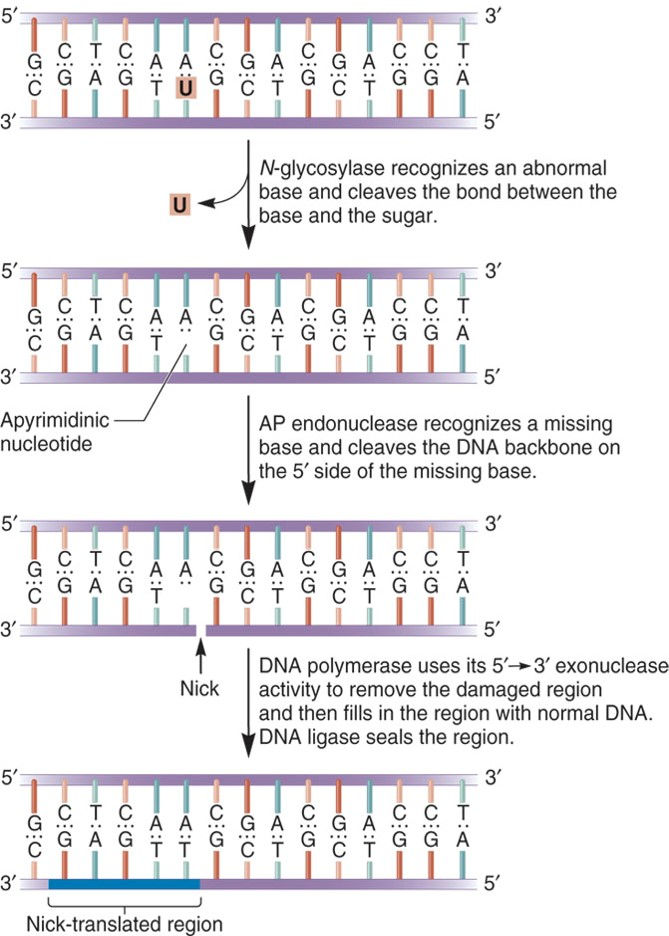
Nucleotide Excision Repair:
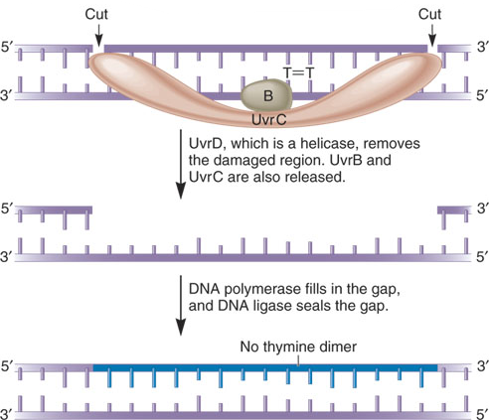
In E. coli, the NER system requires four key proteins:
These are designated UvrA, UvrB, UvrC and UvrD
Named as such because they are involved in Ultraviolet light repair of pyrimidine dimers
They are also important in repairing chemically damaged DNA
UvrA, B, C, and D recognize and remove a short segment of damaged DNA
DNA polymerase and ligase finish the repair job
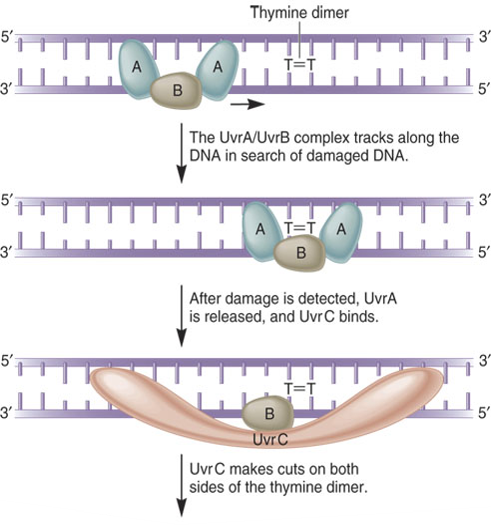
Several human diseases have been shown to involve inherited defects in genes involved in NER
These include:
a)xeroderma pigmentosum (XP)
b)Cockayne syndrome (CS)
A common characteristic of both syndromes is an increased sensitivity to sunlight
Mismatch Repair Systems:
A Base Mismatch is another type of abnormality in DNA.The structure of the DNA double helix obeys the AT/GC rule of base pairing.However, during DNA replication an incorrect base may be added to the growing strand by mistake.DNA polymerases have a 3’ to 5’ proofreading ability that can detect base mismatches and fix them.If proofreading fails, the methyl-directed mismatch repair system comes to the rescue.Mismatch repair systems are found in all species. In humans, mutations in this system are associated with particular types of cancer.
The molecular mechanism of mismatch repair has been studied extensively in E. coli
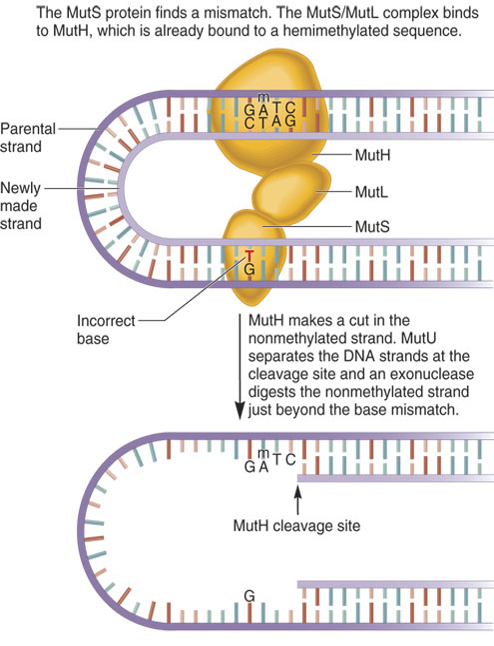
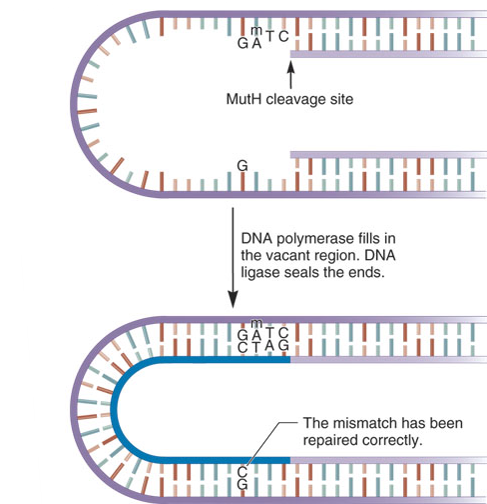
Three proteins are involved named MutL, MutH and MutS
They Detect the mismatch and direct its removal from the newly made strand
The proteins are named Mut because their absence leads to a much higher mutation rate than normal.
A key characteristic of MutH is it can distinguish between the parental strand and the daughter strand prior to replication,both strands are methylated immediately after replication,the parental strand is methylated whereas the daughter is not

Double-Strand Breaks DNA Can Be Repaired by Recombination
DNA double-strand breaks (DSB) are generated in a random manner in the genome by exogenous agents, such as ionizing radiation (IR) or radiomimetic drugs/chemical mutagens. It can also be caused by free radicals which are the byproducts of cellular metabolism.About 10-100 breaks occur each day in a typical human cell,these breaks can cause chromosomal rearrangements and deficiencies. The beauty of DNA repair comes from the structure of DNA;
•The double helix carries two separate copies of all the genetic information in each of its two strands.
• When one strand is damaged, the other is used as a template to restore the sequence to the damaged strand.
•When both strands of the double helix are broken,
–no template strand for repairs, cells may use one of two distinct alternative mechanisms to address breaks.
Two of these systems are known as
1)Non-homologous end joining (NHEJ)
2)Homologous recombination repair (HRR)
Non-Homologous End Joining (NHEJ): It's a quick method for fixing the break.The two broken ends are brought together and joined,usually with the loss of one or two nucleotide letters at the site of joining.Although this loss results in a permanent change in the DNA sequence, it is less harmful for the cell than a persisting double-stranded break.Since very little of our genome actually codes for protein, the resulting mutation is statistically unlikely to be of much consequence.
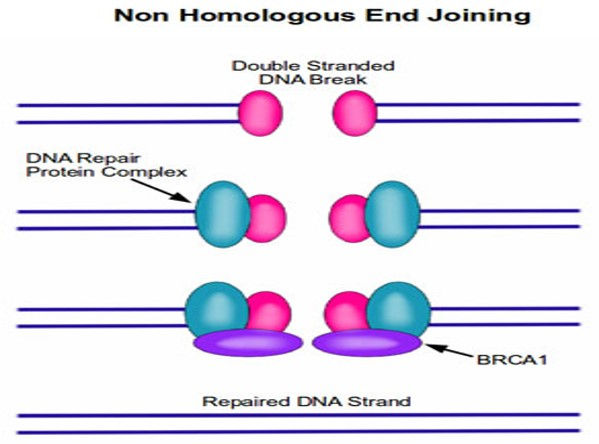
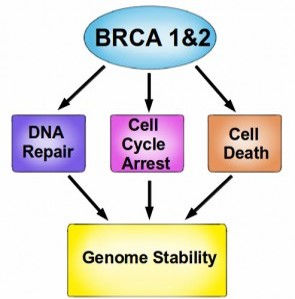
•Two of the most famous proteins in cancer, BRCA1 and BRCA2, play a central role in DNA repair.
•When either BRCA1 or BRCA2 are absent as the result of a mutation, these DNA repair complexes do not form after DNA damage.
•Therefore, cells that are missing BRCA1 or BRCA2 are hypersensitive to agents that damage DNA, such as UV light and various chemical carcinogens.
•For example: Women with an abnormal BRCA1 or BRCA2 gene
–have an extremely high risk of developing breast cancer or developing ovarian cancer.
•As a result, many women who have a family history of breast and ovarian cancers undergo genetic testing.
•If they test positive for either gene mutation, they may opt to undergo a preventative mastectomy.
Homologous Recombination DNA Repair (HRR): HRR Exploits the fact that each cell contains two copies of each double helix (i.e. each cell has two copies of each chromosome).In this case, the repair mechanism is able to transfer nucleotide sequence information from the intact double helix to the broken one.Recombination proteins recognize regions of similarity between the two double helices and bring these regions together.Then, DNA replication proteins use the intact helix as a template to repair the broken one.
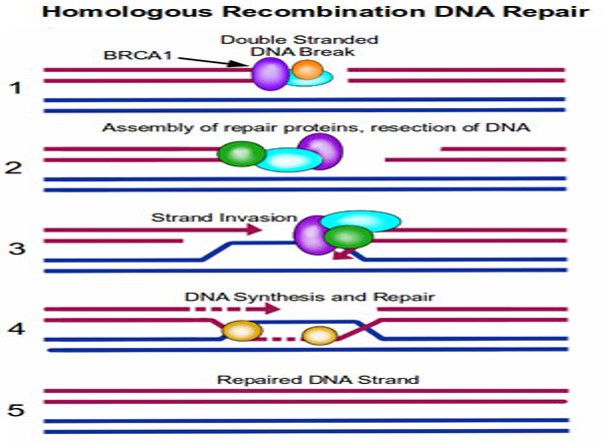
STEP 1. Repair proteins assemble at the site of DNA damage.
STEP 2. The repair proteins resect the DNA to create some room to work in.
STEP 3. The intact copy of DNA (blue strands) then ‘invades’ the damaged strand.
STEP 4. Repair proteins are able to use this intact copy of DNA as a template to repair the break in the damaged strand, by synthesizing DNA across the gap.
STEP 5. The dsDNA break is repaired, with a high level of fidelity.



Comments